ambit2.core.io
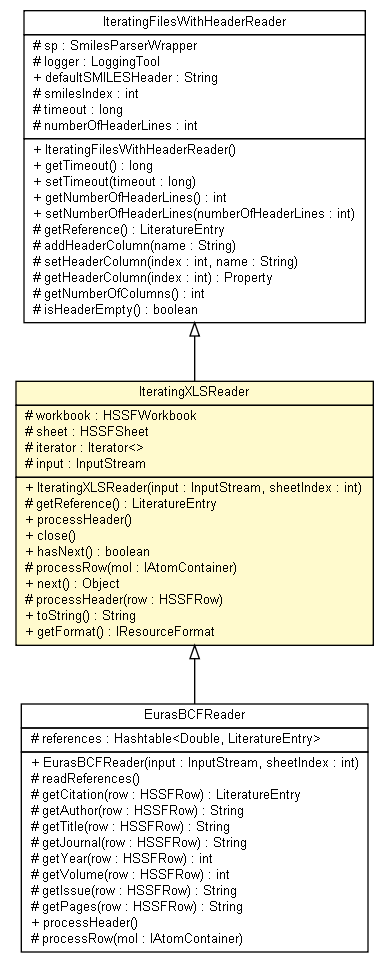
Class IteratingXLSReader
java.lang.Object
 org.openscience.cdk.io.iterator.DefaultIteratingChemObjectReader
org.openscience.cdk.io.iterator.DefaultIteratingChemObjectReader
 ambit2.core.io.IteratingFilesWithHeaderReader
ambit2.core.io.IteratingFilesWithHeaderReader
 ambit2.core.io.IteratingXLSReader
ambit2.core.io.IteratingXLSReader
- All Implemented Interfaces:
- java.util.Iterator, org.openscience.cdk.io.IChemObjectIO, org.openscience.cdk.io.iterator.IIteratingChemObjectReader
- Direct Known Subclasses:
- EurasBCFReader
public class IteratingXLSReader
- extends IteratingFilesWithHeaderReader
Reads XLS files. This implementation loads the workbook in memory which is inefficient for big files.
TODO find how to read it without loading into memory.
|
Field Summary |
protected java.io.InputStream |
input
|
protected java.util.Iterator |
iterator
|
protected org.apache.poi.hssf.usermodel.HSSFSheet |
sheet
|
protected org.apache.poi.hssf.usermodel.HSSFWorkbook |
workbook
|
| Methods inherited from class org.openscience.cdk.io.iterator.DefaultIteratingChemObjectReader |
accepts, addChemObjectIOListener, fireFrameRead, fireIOSettingQuestion, getIOSettings, remove, removeChemObjectIOListener |
| Methods inherited from class java.lang.Object |
clone, equals, finalize, getClass, hashCode, notify, notifyAll, wait, wait, wait |
workbook
protected org.apache.poi.hssf.usermodel.HSSFWorkbook workbook
sheet
protected org.apache.poi.hssf.usermodel.HSSFSheet sheet
iterator
protected java.util.Iterator iterator
input
protected java.io.InputStream input
IteratingXLSReader
public IteratingXLSReader(java.io.InputStream input,
int sheetIndex)
throws ambit2.base.exceptions.AmbitIOException
- Throws:
ambit2.base.exceptions.AmbitIOException
getReference
protected ambit2.base.data.LiteratureEntry getReference()
- Specified by:
getReference in class IteratingFilesWithHeaderReader
processHeader
public void processHeader()
close
public void close()
throws java.io.IOException
- Throws:
java.io.IOException
hasNext
public boolean hasNext()
processRow
protected void processRow(org.openscience.cdk.interfaces.IAtomContainer mol)
next
public java.lang.Object next()
processHeader
protected void processHeader(org.apache.poi.hssf.usermodel.HSSFRow row)
toString
public java.lang.String toString()
- Overrides:
toString in class java.lang.Object
getFormat
public org.openscience.cdk.io.formats.IResourceFormat getFormat()

org.openscience.cdk.io.iterator.DefaultIteratingChemObjectReader
ambit2.core.io.IteratingFilesWithHeaderReader
ambit2.core.io.IteratingXLSReader