ambit2.core.io
Class MDLV2000ReaderExtended
java.lang.Object
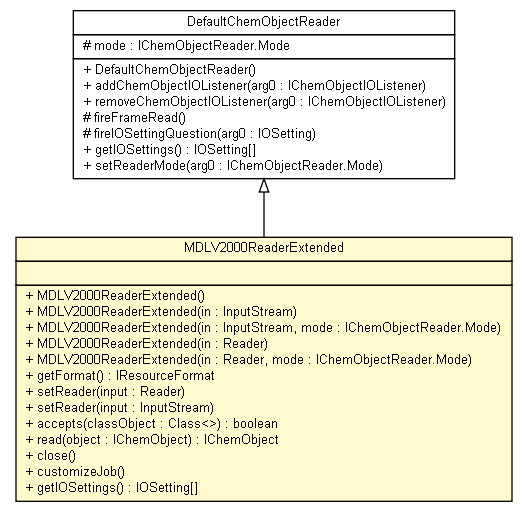
 org.openscience.cdk.io.DefaultChemObjectReader
org.openscience.cdk.io.DefaultChemObjectReader
 ambit2.core.io.MDLV2000ReaderExtended
ambit2.core.io.MDLV2000ReaderExtended
- All Implemented Interfaces:
- org.openscience.cdk.io.IChemObjectIO, org.openscience.cdk.io.IChemObjectReader
public class MDLV2000ReaderExtended
- extends org.openscience.cdk.io.DefaultChemObjectReader
Supports SGroups.
Reads a molecule from an MDL MOL or SDF file {cdk.cite DAL92}. An SD files
is read into a ChemSequence of ChemModel's. Each ChemModel will contain one
Molecule.
From the Atom block it reads atomic coordinates, element types and
formal charges. From the Bond block it reads the bonds and the orders.
Additionally, it reads 'M CHG', 'G ', 'M RAD' and 'M ISO' lines from the
property block.
If all z coordinates are 0.0, then the xy coordinates are taken as
2D, otherwise the coordinates are read as 3D.
The title of the MOL file is read and can be retrieved with:
molecule.getProperty(CDKConstants.TITLE);
RGroups which are saved in the mdl file as R#, are renamed according to their appearance,
e.g. the first R# is named R1. With PseudAtom.getLabel() "R1" is returned (instead of R#).
This is introduced due to the SAR table generation procedure of Scitegics PipelinePilot.
cdk.module io
cdk.svnrev $Revision: 9684 $
| Nested classes/interfaces inherited from interface org.openscience.cdk.io.IChemObjectReader |
org.openscience.cdk.io.IChemObjectReader.Mode |
| Fields inherited from class org.openscience.cdk.io.DefaultChemObjectReader |
mode |
|
Method Summary |
boolean |
accepts(java.lang.Class classObject)
|
void |
close()
|
void |
customizeJob()
|
org.openscience.cdk.io.formats.IResourceFormat |
getFormat()
|
org.openscience.cdk.io.setting.IOSetting[] |
getIOSettings()
|
org.openscience.cdk.interfaces.IChemObject |
read(org.openscience.cdk.interfaces.IChemObject object)
Takes an object which subclasses IChemObject, e.g. |
void |
setReader(java.io.InputStream input)
|
void |
setReader(java.io.Reader input)
|
| Methods inherited from class org.openscience.cdk.io.DefaultChemObjectReader |
addChemObjectIOListener, fireFrameRead, fireIOSettingQuestion, removeChemObjectIOListener, setReaderMode |
| Methods inherited from class java.lang.Object |
clone, equals, finalize, getClass, hashCode, notify, notifyAll, toString, wait, wait, wait |
MDLV2000ReaderExtended
public MDLV2000ReaderExtended()
MDLV2000ReaderExtended
public MDLV2000ReaderExtended(java.io.InputStream in)
- Contructs a new MDLReader that can read Molecule from a given InputStream.
- Parameters:
in - The InputStream to read from
MDLV2000ReaderExtended
public MDLV2000ReaderExtended(java.io.InputStream in,
org.openscience.cdk.io.IChemObjectReader.Mode mode)
MDLV2000ReaderExtended
public MDLV2000ReaderExtended(java.io.Reader in)
- Contructs a new MDLReader that can read Molecule from a given Reader.
- Parameters:
in - The Reader to read from
MDLV2000ReaderExtended
public MDLV2000ReaderExtended(java.io.Reader in,
org.openscience.cdk.io.IChemObjectReader.Mode mode)
getFormat
public org.openscience.cdk.io.formats.IResourceFormat getFormat()
setReader
public void setReader(java.io.Reader input)
throws org.openscience.cdk.exception.CDKException
- Throws:
org.openscience.cdk.exception.CDKException
setReader
public void setReader(java.io.InputStream input)
throws org.openscience.cdk.exception.CDKException
- Throws:
org.openscience.cdk.exception.CDKException
accepts
public boolean accepts(java.lang.Class classObject)
read
public org.openscience.cdk.interfaces.IChemObject read(org.openscience.cdk.interfaces.IChemObject object)
throws org.openscience.cdk.exception.CDKException
- Takes an object which subclasses IChemObject, e.g. Molecule, and will read
this (from file, database, internet etc). If the specific implementation
does not support a specific IChemObject it will throw an Exception.
- Parameters:
object - The object that subclasses
IChemObject
- Returns:
- The IChemObject read
- Throws:
org.openscience.cdk.exception.CDKException
close
public void close()
throws java.io.IOException
- Throws:
java.io.IOException
customizeJob
public void customizeJob()
getIOSettings
public org.openscience.cdk.io.setting.IOSetting[] getIOSettings()
- Specified by:
getIOSettings in interface org.openscience.cdk.io.IChemObjectIO- Overrides:
getIOSettings in class org.openscience.cdk.io.DefaultChemObjectReader

org.openscience.cdk.io.DefaultChemObjectReader
ambit2.core.io.MDLV2000ReaderExtended